Research Overview⌗
In the Walker group, we are interested in how to connect biological phenomena and measurements of fluorescence using computational chemistry. Molecular modeling provides ways of translating between what we see in experiment, usually a signal or set of signals, to atomic or electronic level details of the system. To investigate fluorescence in biological systems, we use and develop new techniques in quantum chemistry, molecular dynamics, and excited state simulations. This has direct applications in imaging, both in medicine and in basic science.
Fluorescent Nucleotides and Aptamers⌗
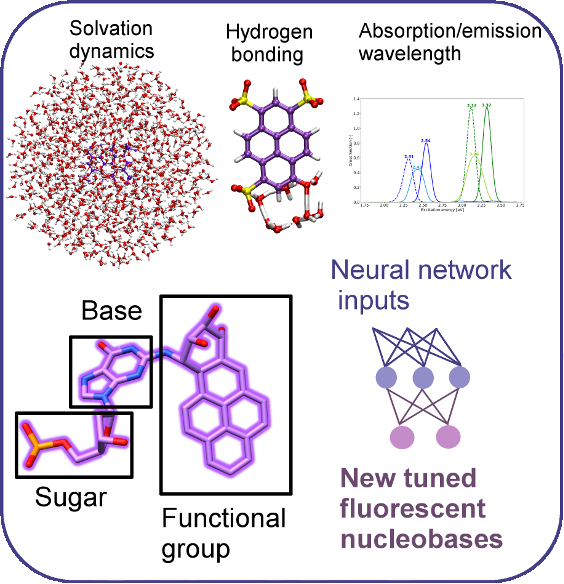
Theoretical chemistry provides avenues of testing large numbers of compounds and hypotheses in a systematic and inexpensive way, and also allows us to uncover principles for further design. We work to design and understand complex biomolecular scaffolds for drug design, including designing new fluorescent biomolecules and sugar-based drug targets with multiscale modeling and machine learning. Supported by R35GM154949.
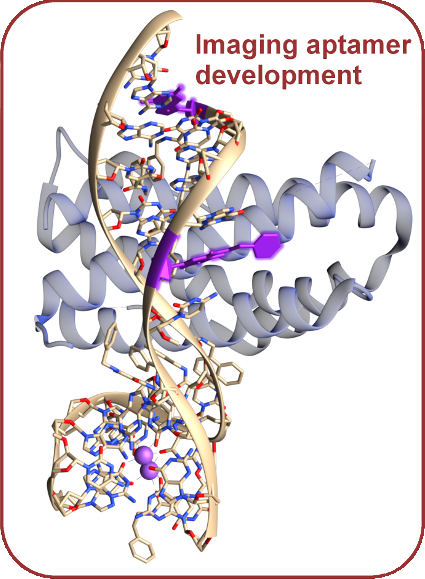
Aptamers, or short sequences of nucleotides, are easy to synthesize and are highly specific to their targets. By modifying specific parts of existing tight-binding aptamer drugs, we can potentially predict functionalization locations and effects of adding fluorescent synthetic nucleotides to various points in existing aptamer drugs, allowing for potential control and improved interpretation of experimental signals.
Fluorescent Protein Design⌗
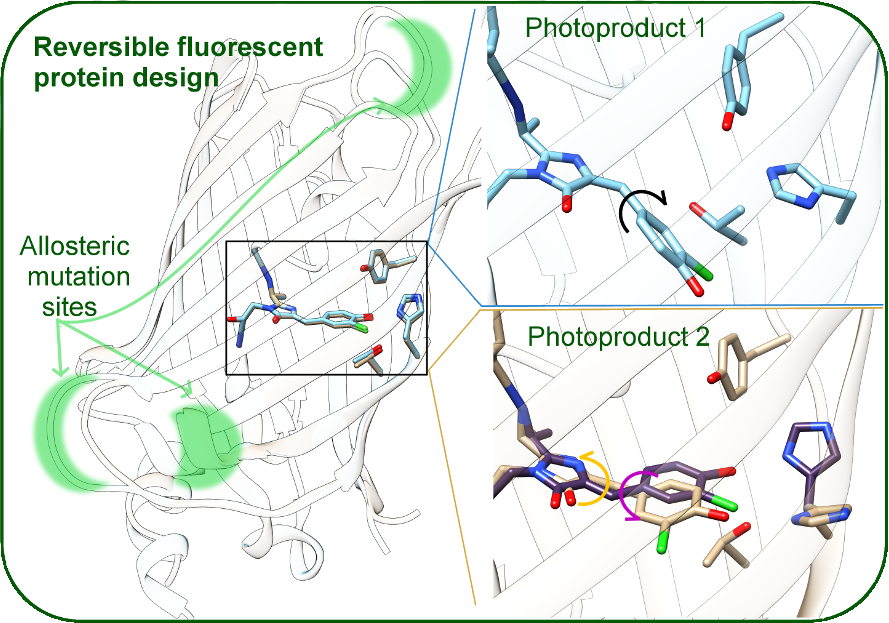
Fluorescent proteins and their derivatives are powerful experimental tools. Our group applies classical and combined quantum/molecular mechanics methods on the ground and excited state for the rational design of fluorescent proteins (FPs), based on a comprehensive theoretical picture of the relationship between excited state chromophore behavior, protein structure, and environment. Fluorescent proteins are ubiquitous as sensors in biochemical imaging, but the relationship between protein structures and motion, environmental factors such as pH, chromophore excited state behavior, and the actual measured fluorescent signal is still not fully understood. Since the connection between targeted mutations and experimental spectroscopic outcomes is not predictable, designing new FP-based sensors is an ongoing challenge. We are especially interested in specific challenges associated with creating bright, red, monomeric or photodissociative sensors for specific anions of interest, combining theoretical methodology and experimental collaborations. Our work will also give further insight into the connection between atomic and electronic-level details of protein structure and measured spectroscopic signals. Supported by NSF CHE2338804.
FRET Tag Simulations of RNA Pol II⌗

Many experiments use fluorescent tags to investigate protein dynamics. We investigate large protein systems with their fluorescent tags attached, both to compare fluorescent emission wavelengths and brightness to experimental quantities and to potentially design custom tags with new properties. As the tags become close to each other, energy transfers from Tag 1 to Tag 2, resulting in an immediate shift in emission wavelength. This can be used to compare the motions of proteins simulated on the computer with the motions obtained in experiment, while also providing greater detail on what is happening on the atomic level for even very large systems.